Quality Control
One of the most commonly used tools for the basic quality control of the raw reads in fastq format and, at the same time, probably as easy-to-use as it gets, since the command to run it looks like this:
fastqc <filename.fastq>
The results are given in the .html file, which you have to download to your desktop and view in the webbrowser, as we cannot view .html files directly at the server (for now). The scheme of their presentation is quite beginner-friendly - everything is presented visually (on graphs & tables), so there’s no need to browse through countless lines of text to retrieve the information you need
It should look more or less like that:
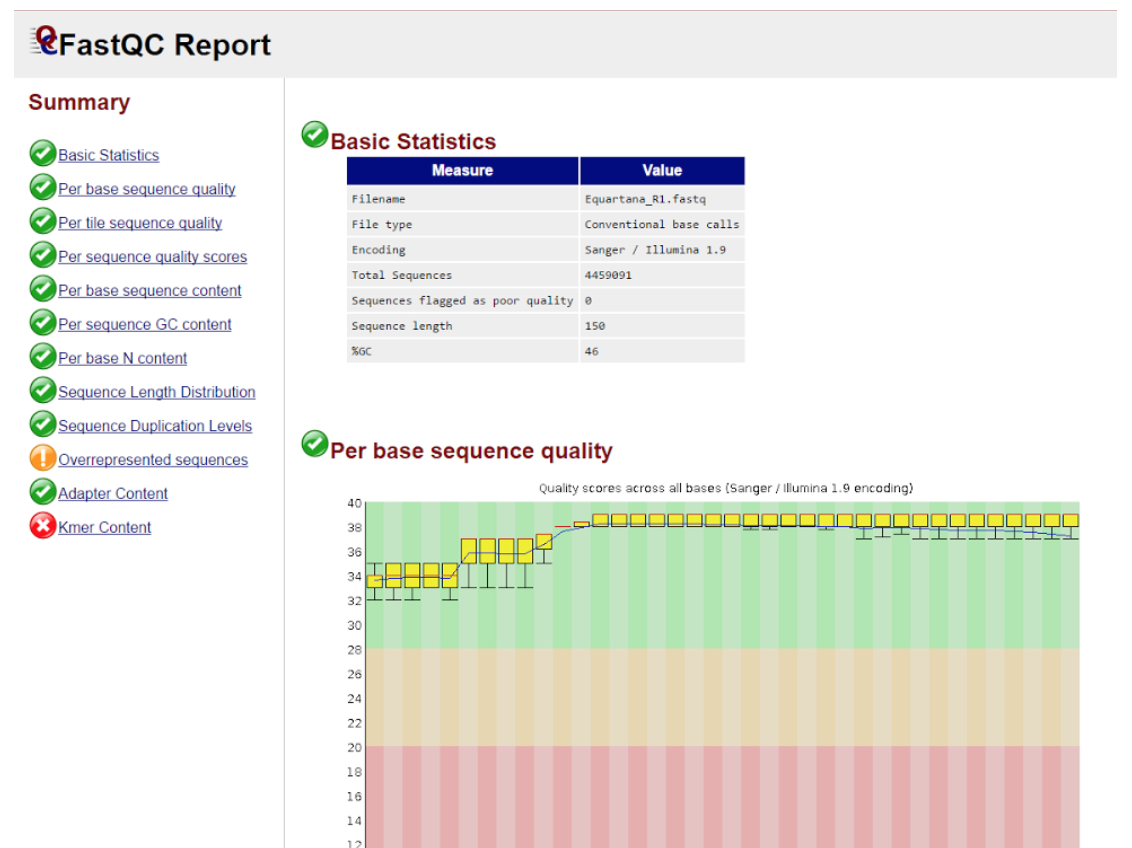
Most “chapters” of the results are rather self-explanatory, but just in case the vocabulary used there does not seem so, here’s a short guide:
SCORES are given as Phred quality values, which are logarithmic representations of error probability (more here). Technically speaking, they could go up to infinity (as does a logarithmic function); however, in fastqc there’s a clear cutoff on the value of 40 - this is because a score of 40 is equal to error probability of 1 in 10 000, which in practice translates to “as good as it could ever be”.
TILES are physical locations on the matrix inside the Illumina sequencing machine. Quality per tile is most likely not informative in any way for the user, but a very poor score for a specific tile in consecutive runs of the sequencer might indicate a hardware fault, which would be worth reporting to the machine’s owner.
N in “per base N content” stands for an unknown nucleotide - any of the regular four - which basically means that the sequencer couldn’t identify it with full certainty. There are countless reasons why this occurs and as long as N’s do not make up a significant part of the sequence, it’s not something you should worry about.
OVERREPRESENTED SEQUENCES are, well, obviously sequences that occur disproportionately often in the obtained data. What might not be obvious is what it means in practice - usually you will notice only one overrepresented sequence (or none at all), and most probably it’s going to be the sequence of the adapter. What an adapter is and how to deal with it will be explained in the following chapters.